The Landeira lab is driven by the belief that many aspects of human health are not genetically predetermined. We hypothesize that the onset of aging-associated diseases is partly a consequence of the deterioration of the cellular epigenetic memory, eventually leading to changes in cell identity and the onset of aging-associated diseases including cancer (Figure 1). We aim to contribute to establish the molecular foundation of epigenetic memory, to identify which are the key epigenetic pathways that drive aging and cancer progression, and to the design of new therapies that reverse aging-associated diseases. We are currently developing three research lines in the lab:

Figure 1. Scheme summarizing the general research interest of the Landeira lab in epigenetics during embryo development and in human disease.
What is the molecular basis of epigenetic memory?
To treat epigenetic-based diseases we first need to establish the key molecular epigenetic pathways that facilitate the formation and functioning of complex multicellular organisms like humans. Upon fusion of the sperm and oocyte cells, a single cell named the zygote is formed. The zygote has a unique genome and during organism development it goes through a vast number of proliferation and differentiation rounds that generate over 1012 cells that are genetically identical but functionally distinct, and that self-organise to produce 78 organs that compose the adult human body. Cell specialization depends on the establishment of distinct epigenetic configurations that facilitate the expression of set of cell-type-specific genes during embryo development. Thus, cell-type-specific epigenetic patterns need to be “memorized” and maintained for a lifetime, because the original differentiation signal instructing the change in gene expression during prenatal life is no longer present during the adult life. Importantly, different types of environmental stimulus can perturb the cells’ epigenetic memory, and thus we hypothesize that many diseases associated to aging are a consequence of the perturbation of the original epigenetic configuration that was laid on the genome during development. In our lab we are dissecting the mechanism by which epigenetic regulators promote the maintenance of epigenetic memory. In particular, we study the molecular details as to how two hallmark epigenetic systems (Polycomb proteins and H3K9 methyltransferases) enable the perpetuation of transcriptional memory in mouse pluripotent stem cells. Our recent findings (1,2) suggest that cellular memory is supported by an epigenetic cycle in which reversible activities carried out by epigenetic regulators in coordination with cell cycle transition create a multiphasic system that can accommodate both maintenance of cell identity and cell differentiation in proliferating stem cell populations (3) (Figure 2).
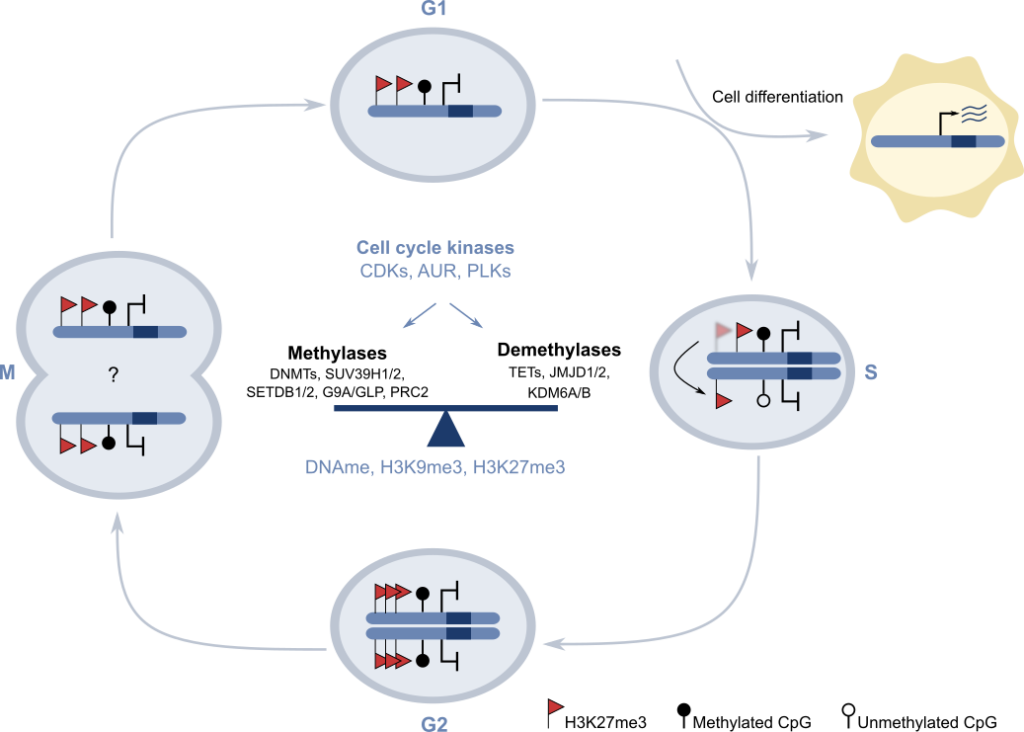
Figure 2. Model proposing the molecular basis of epigenetic memory: an “Epigenetic cycle” in which epigenetic regulators are coordinated with cell kinases during cell cycle transition to allow both self-renewal and cell differentiation in stem cells. Adapted from Espinosa-Martinez et al. Science Advances 2020.
Is the loss of epigenetic information the reason we age? Can we slow down the pace of aging using drugs that stabilize our epigenome?
Deciphering the molecular basis of human aging is an exciting challenge in fundamental biology with great implications in public health. Emerging reports suggest that aging can be induced by environmental factors that challenge the stability of epigenetic information stored on our genomes. We support the idea that the aging process is driven by the progressive loss of “youthful epigenetic information” laid during development, the retrieval of which via epigenetic reprogramming can improve the function of damaged and aged tissues by catalysing age reversal (4) (Figure 3). Using pluripotent cells and mutant mice as model systems, we are currently investigating which are the epigenetic factors that constitute our young epigenome, and that protects us from aging during lifespan. For example, building on a previous report showing that the function of the molecular clock protein BMAL1 during development is critical to avoid early aging in adult mice (5), we have found that in pluripotent cells BMAL1 is redeployed as transcriptional repressor of transposable elements through H3K9me3 independently of CLOCK (6), highlighting that epigenetic perturbations during prenatal life can have a drastic impact in mammalian health and life span. We envision that characterization of such mechanisms will provide a better understanding of the molecular basis of aging and allow us to devise safe pharmacological interventions that protect us from the loss of youth-encoding epigenetic memory during our lifespan.

Figure 3. Scheme showing the epigenetic landscape of development, aging and rejuvenation. Inspired on the original Waddington metaphor, valleys represent more stable states of cell identity that are separated with each others by walls (epigenome) during developmental specification. As we age, walls are perturbed and cell identity becomes less stably determined. Reprogramming strategies favouring reestablishment of the epigenome are indicated. Adapted from Ryan Lu et al. Nature Aging 2023.
Is epigenetic instability a driver of cancer progression? Can we cure cancer by modulating the cancer epigenome?
Cancer is one of the most prevalent diseases in aged populations. Metastasis is the cause of ~90% of cancer-associated mortality and therefore, to cure cancer, it is urgent to develop new therapies that block the capacity of cancer cells to disseminate throughout the human body. Metastatic dissemination relies on the continuous positive selection of cancer cells that are functionally optimized to survive in the different physiological and pharmacological environments occurring during disease progression. In our lab, we believe that the functional cell-to-cell heterogeneity that drives cancer progression relies, not only on different combination of genetic mutations accumulated in their genome, but also on distinct epigenetic configurations that dictate the behaviour of individual cells (7). We use cancer cells derived from human patients and mouse xenotransplant models to investigate how epigenetic memory affects cancer progression, with the aim to contribute to the development of new therapies that block disease progression by targeting epigenetic regulators. We have recently demonstrated that the epigenetic proteins Polycomb enable phenotypic epithelial-mesenchymal plasticity in cancer cells that facilitate metastatic dissemination (8,9) (Figure 4). These findings provide key information for effective application of currently available Polycomb inhibitors in human cancer patients.

Figure 4. Regulation of metastasis by the epigenetic regulator EZH2. A) Scheme summarizing the role of the Polycomb protein EZH2 in regulating epithelial to mesenchymal transition during carcinoma metastasis. B) Immunohistochemistry pictures of tumors formed in the lungs of mice demonstrating that lung cancer cells require EZH2 for efficient tumor colonization. Adapted from Gallardo et al. Oncogene 2023.
Selected bibliography
1. H. G. Asenjo, M. Alcazar-Fabra, M. Espinosa-Martínez, L. Lopez-Onieva, A. Gallardo, E. Dimitrova, A. Feldmann, T. Pachano, J. Martorell-Marugán, P. Carmona-Sáez, A. Sanchez-Pozo, Á. Rada-Iglesias, R. J. Klose, D. Landeira, Changes in PRC1 activity during interphase modulate lineage transition in pluripotent cells. Nat Commun 14, 180 (2023).
2. H. G. Asenjo, A. Gallardo, L. López-Onieva, I. Tejada, J. Martorell-Marugán, P. Carmona-Sáez, D. Landeira, Polycomb regulation is coupled to cell cycle transition in pluripotent stem cells. Science advances 6, eaay4768 (2020).
3. M. Espinosa-Martínez, M. Alcázar-Fabra, D. Landeira, The molecular basis of cell memory in mammals: The epigenetic cycle. Science advances 10, eadl3188 (2024).
4. Y. R. Lu, X. Tian, D. A. Sinclair, The Information Theory of Aging. Nature aging 3, 1486-1499 (2023).
5. G. Yang, L. Chen, G. R. Grant, G. Paschos, W. L. Song, E. S. Musiek, V. Lee, S. C. McLoughlin, T. Grosser, G. Cotsarelis, G. A. FitzGerald, Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Science translational medicine 8, 324ra316 (2016).
6. A. Gallardo, E. Belmonte-Reche, M. Marti-Marimon, J. Domingo-Reines, G. Peris, L. Lopez-Onieva, I. Fernandez-Rengel, P. Tristan-Ramos, N. Bellora, A. Sanchez-Pozo, A. M. Estevez, S. R. Heras, M. Marti-Renom, D. Landeira, BMAL1 represses transposable elements independently of CLOCK in pluripotent cells. bioRxiv, 2024.2006.2018.599568 (2024).
7. W. A. Flavahan, E. Gaskell, B. E. Bernstein, Epigenetic plasticity and the hallmarks of cancer. Science 357, (2017).
8. A. Gallardo, L. López-Onieva, E. Belmonte-Reche, I. Fernández-Rengel, A. Serrano-Prados, A. Molina, A. Sánchez-Pozo, D. Landeira, EZH2 represses mesenchymal genes and upholds the epithelial state of breast carcinoma cells. Cell death & disease 15, 609 (2024).
9. A. Gallardo, A. Molina, H. G. Asenjo, L. Lopez-Onieva, J. Martorell-Marugán, M. Espinosa-Martinez, C. Griñan-Lison, J. C. Alvarez-Perez, F. E. Cara, S. A. Navarro-Marchal, P. Carmona-Sáez, P. P. Medina, J. A. Marchal, S. Granados-Principal, A. Sánchez-Pozo, D. Landeira, EZH2 endorses cell plasticity to non-small cell lung cancer cells facilitating mesenchymal to epithelial transition and tumour colonization. Oncogene 41, 3611-3624 (2022).